Summary of microscopy work
Read on for some background, or jump ahead to the discussion of my contributions.
Background
The starting point for my work was Richardson-Lucy deconvolution, a deblurring technique invented almost fifty years ago. The algorithm is mechanically very simple. Each iteration is a multiplicative update of the previous guess, where the update depends on the ratio between the measured image and the expected image generated by the current guess:
# H is a linear operator mapping estimates to images
# H_T is the transpose of H (and thus maps images to estimates)
def richardson_lucy(img, guess=None, n_iter=64):
if guess is None:
guess = np.ones_like(H_T(img))
normalizer = H_T(np.ones_like(img))
for _ in range(n_iter):
guess *= H_T(img / H(guess)) / normalizer
return guess
Richardson-Lucy corresponds to a special form of gradient descent to find the maximum likelihood estimate assuming that each pixel of the image is corrupted by Poisson noise. The distinction from usual gradient descent is that the step size is a fixed scalar multiple of the current estimate. One nice feature of the multiplicative update is that (given a feasible estimate and a positive semidefinite blurring operator $\mathbf{H}$) it automatically preserves a non-negativity constraint on the estimate. Much of the performance of Richardson-Lucy derives from this constraint.
The technique is called deconvolution because the blurring induced by $\mathbf{H}$ is represented as convolution with some point spread function (the image of a single point). In most deconvolution literature, $\mathbf{H}$ is a pure convolution. However, the algorithm works just as well for any linear operator $\mathbf{H}$. It is best suited for linear operators with special structure that allow fast computation; this usually means that $\mathbf{H}$ is diagonal either directly or following a Fourier transform. Ingaramo et al. point out that a broad range of computational microscopy tasks can be attacked by approximately inverting some linear $\mathbf{H}$. In particular, techniques that combine multiple images (which could come from multiple viewing angles, or different illumination conditions) to arrive at a final estimate of the object can be described this way. This makes Richardson-Lucy a versatile tool for image reconstructions.
Inverse problems — solving for an unknown ground truth given data and a known forward model — are usually ill-posed. The loss of information in mapping from object to image often means that many solutions are consistent with the data. The classic solution to this issue is regularization, where an additional penalty function is added to distinguish among solutions. This is equivalent to imposing a prior on your object space. The penalty function says that solutions with certain characteristics are less probable than others. Many classic regularization methods (e.g. Tikhonov, total variation) have sensible interpretations and can be made to work well. However, there often aren’t good ways to decide on how to parameterize them, leaving non-experts stuck trying to choose parameters instead of having a tool that “just works”. The tweakability of these parameters also leads (particularly in the biological microscopy community) to concerns about adjusting them until you “see what you want to see”.
In Andrew York’s lab, we adopted a different approach. Instead of using priors on the structure of the object, we decided to pursue priors based on the photophysical behavior of the molecules being imaged. This has nice conceptual advantages over other approaches. (And, if you like some of the usual regularization priors, you can still include them in our enlarged models.) Our models mostly depend on measurable parameters, resolving the parameterization criticisms of the previous paragraph. This approach also offers a way to tie-in to single molecule localization microscopy, one of the great microscopy ideas of recent times.
Localization microscopy exploits the enormous gain in precision offered by asking “how precisely can I locate a single emitter?” instead of “how precisely can I resolve an unknown number of emitters?” With the right choice of fluorophores, one can engineer situations where very few emitters are active at any given time. If the emitters are sufficiently isolated in each frame, this lets you decompose your image into a sequence of single particle localizations. When this works, it yields an image with resolution far better than the usual diffraction limit, highlighting the power of photophysical priors. However, it poses difficult problems in sample preparation, and when the fluorophores are not sufficiently well isolated, standard reconstruction algorithms can produce serious distortions. An algorithm that, using the available data where it can, moves seamlessly between the sparsely tagged regime of localization microscopy and the densely tagged regime of widefield imagery could be profoundly influential.
We believe that combining deconvolution with photophysics is a natural road to that goal. The recent explosion of work applying neural networks to microscopy image problems suggests another worthy approach. While I’ve experimented some with these techniques, we feel there are advantages to the maximum likelihood approach that make it worth studying. Chief among them is that in our approach, there is a clean separation between properties of your tools (the fluorophores and imaging system) and your sample. With neural networks, the reconstructions will usually depend on both the imaging system and the samples you’ve used for training. Having a distinction between properties of the model and properties of the sample is conceptually satisfying from an interpretability standpoint. It also suggests an easier road to generalizing, either to different types of biological samples or to multiple realizations of an instrument.
My work
My first project incorporated photobleaching into a deconvolution algorithm. Photobleaching is a process where emitting molecules irreversibly stop glowing. We chose it as a first effect to add because photobleaching is both ubiquitous and easily modeled. To a good approximation, over short times the conditional probability of $x_{t+1}$ emitters remaining unbleached at time $t+1$ given $x_{t}$ at time $t$ is binomially distributed. The probability of bleaching depends on the illumination dose received during each exposure. Typically we assume uniform illumination, corresponding to well-executed widefield microscopy, but the method can be tailored to other situations.
Instead of a single exposure, our algorithm considers a time series of images of a fixed sample. This lets us measure photobleaching information to include in the reconstruction. We then use a joint likelihood function for all the frames, combining the Poisson likelihood associated with the images with the binomial likelihood associated with photobleaching. To get around the discreteness of the binomial, we use an interpolated version where factorials are replaced by gamma functions. Because gamma functions continue to be well-defined for negative real numbers, we must explicitly enforce a non-increasing constraint over time at each pixel to reflect the monotonic nature of bleaching. Richardson-Lucy is unequipped to handle this constraint, so initially we used projected gradient descent, with the pool adjacent violators algorithm (PAVA) providing a fast projector. Later, we realized that you can also work in a “decay basis” where the estimate is expressed as the number of bleaching events that occur between one frame and the next. In this basis, you once again have a simple non-negativity constraint.
Tests on synthetic data showed that we achieved many of our goals. The
new method can run for many more iterations (and closer to
convergence) than Richardson-Lucy before showing high spatial
frequency artifacts, demonstrating the regularizing effect of the
extra information. Intriguingly, in appropriate conditions our
algorithm also demonstrates a capacity for localization. The figure
below shows how this arises. At late time points —
corresponding to moving down the y-axis on each panel — many of the
emitters have bleached out, leaving a very sparse distribution. In
this situation Poisson maximum likelihood does very well as a
localization algorithm, and the non-increasing constraint imposed by
photobleaching gives us a way to propagate this information back to
earlier time points. 
Further testing revealed one unpleasant bias of the algorithm. For combinatorial reasons, the bleaching part of the likelihood favors solutions where density is concentrated in a single pixel rather than spread over multiple pixels. In situations where the spatial information isn’t strong enough to counteract this — a spatially oversampled image, for example — overly compact solutions are favored. By exactly solving a restricted version of the problem using the forward-backward algorithm for hidden Markov models, I showed this arises from looking at the maximum likelihood for the full trajectory rather than the marginal probability distribution of emitters at the first time point. To counteract this problem, we experimented with a different loss function that penalizes bleaching trajectories that are far from the expected exponential decay. This does combat the clumping effect, but in the restricted problem one finds it now biases toward “anti-clumped” data. Trying to find the right approximation to the marginal at the first time point (or using MCMC techniques to calculate it) remains an open problem.
The photobleaching deconvolution project led to interesting related work: understanding the optimal exposure time given photoswitching. I became interested in this question while testing deconvolution with photobleaching. Our method takes a time series of images; one knob this gives you is how many slices you want to take for a fixed total exposure time. Since Poisson random variables are stable under addition, you can recombine the slices to retrieve the full exposure without distorting statistics. So, smaller time slices give extra temporal information without affecting the total Poisson information available at each pixel. (In practice, non-Poisson contributions to noise would start to become important at short exposures, but in our idealization we don’t have this problem.)
Basic Richardson-Lucy, on the other hand, lacks a way to integrate multiple frames and thus loses signal as we divide time more finely. Comparing our algorithm to the results of Richardson-Lucy performed on just the first frame is unfair. By exploiting the Poisson stability, we can make a synthetic single exposure by summing multiple frames. However, it’s not clear what the correct number of frames (and thus total exposure time) should be. With photobleaching, longer exposure times lower the relative amount of Poisson noise but increase the importance of fluctuations in the time each emitter survives before bleaching. When we checked the microscopy literature on this issue, we didn’t find anything. Given the ubiquity of photobleaching and the simplicity of the question, this was a surprising gap and a pleasant opportunity.
I developed theory for determining optimal exposure times in the
presence of photophysical noise. In the case of single exponential
photobleaching — the same model we considered for our deconvolution work —
I was able to derive limiting results.
The theory also generalizes to other forms of photophysical noise where you
must instead measure the variance in the emitter fluctuations. We think one
reason this noise went unremarked on in the field for a long time is because
it has a very different character from Poisson noise. Photophysical noise
is blurred by the microscope’s point spread function. Consequently
images dominated by photobleaching noise look smooth on a pixel-to-pixel level.
This is very different from how Poisson noise from low photodose looks, and
so bleaching noise distortions can easily be attributed to nonuniformities of
the sample or its labeling:
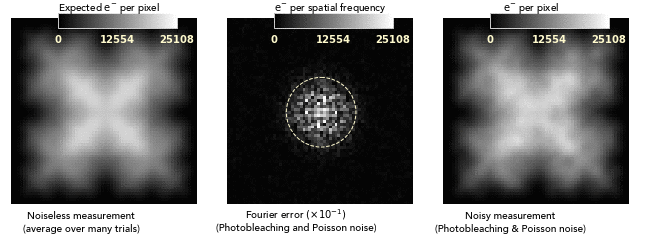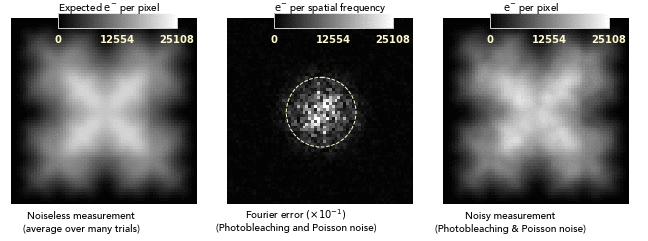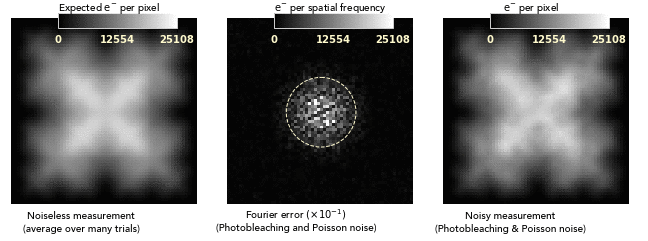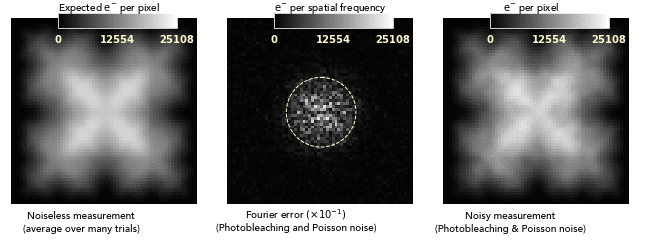 Another consequence of the noise blurring is that the balance between
Poisson and photobleaching noise varies depending on spatial
frequency. Thus, a single exposure time is not well suited for
capturing all features of the image. One must either pick a compromise
time that does best across the feature band of interest, or use
multiple exposures (using the summing trick) and fuse the best
estimates at each spatial frequency. I developed both approaches in
our lab.
Another consequence of the noise blurring is that the balance between
Poisson and photobleaching noise varies depending on spatial
frequency. Thus, a single exposure time is not well suited for
capturing all features of the image. One must either pick a compromise
time that does best across the feature band of interest, or use
multiple exposures (using the summing trick) and fuse the best
estimates at each spatial frequency. I developed both approaches in
our lab.
A key parameter for this model is the expected number of counts generated by an emitter before bleaching. A surprising result of our work is that increasing this value has less impact on the signal to noise ratio than one might expect. A well known property of Poisson noise is that with $N$ counts, the signal to noise ratio (SNR) is $N^{1/2}$. If we let $\langle N \rangle$ denote the expected counts before bleaching, then we’d expect that SNR should scale like $\langle N \rangle^{1/2}$. But, when photobleaching noise is taken into account, the SNR scales as $\langle N \rangle^{1/4}$ at optimal exposure. You can’t use all of the extra signal that you get because it leads to excess photobleaching noise. One consequence of this is that you’re usually better off improving signal by using a larger number of fast-bleaching tags rather than fewer slow-bleaching tags.
For more details on our exposure time theory, please see the slides from our talk at the 2019 Focus on Microscopy conference.
Another project I worked on was extending Richardson-Lucy to deal with nonlinear image formation processes. Many useful sorts of photophysics depend on interactions between molecules. These can’t be captured by a linear imaging process. The main motivating example is FRET, a form of coupling where one “donor” fluorophore can be excited and then give its energy to another “acceptor” emitter. This only occurs when the fluorophores are within a few nanometers of each other, well below the effective point spread function even of superresolution techniques like STED. Examining what sorts of information become available if you include constraints of this precision is exciting.
The modifications to Richardson-Lucy to allow nonlinear image formation end up being quite simple. You can decompose the image formation operator $\mathbf{H}$ into a sequence of linear and nonlinear operations: \begin{equation} \mathbf{H} = \mathbf{H_1} \mathbf{H_2} \ldots \mathbf{H_n}. \nonumber \end{equation} Richardson-Lucy uses the transpose of $\mathbf{H}$, which is easily constructed given linear constituents: \begin{equation} \mathbf{H}^T = \mathbf{H_n}^T \ldots \mathbf{H_2}^T \mathbf{H_1}^T \nonumber \end{equation} If $\mathbf{H}$ contains nonlinearities, the same trick works, provided that when we have nonlinear components we use the transpose of the Jacobian of that component. For some nonlinearities, this violates the assumption that $\mathbf{H}^T$ is positive semidefinite. To maintain Richardson-Lucy’s constraint preserving property in this case, we simply rescale the step size as needed to avoid going negative anywhere.
To use nonlinear Richardson-Lucy to test the possibilities of joint FRET reconstructions with deconvolution, I developed an efficiently computable model of imaging with FRET. I validated the nonlinear Richardson-Lucy algorithm by comparing it to other optimization algorithms on the same loss function. Testing nonlinear Richardson-Lucy on synthetic FRET data, it is clear that further progress on this topic requires other forms of regularization. This is unsurprising; to really capture the possibilities of FRET interactions requires either a heavily oversampled image or reconstruction on a much smaller pixel grid used in the image. The additional loss of information presumably results in a larger manifold of equivalent seeming solutions with regard to the loss function. One possibility is to incorporate other techniques (like our photobleaching work) to provide more information, although the computational demands become formidable. Another possibility is to look at engineering longer range interactions between emitters to exploit.